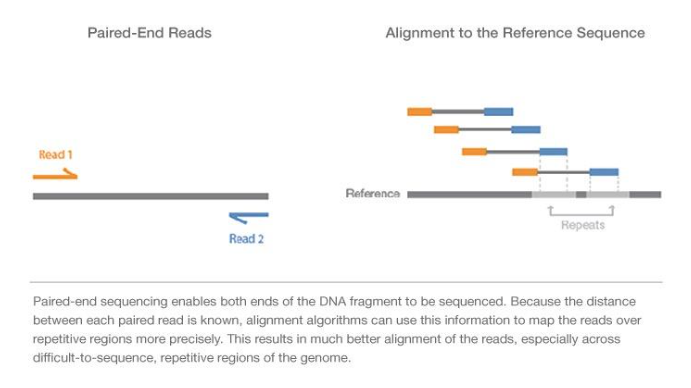
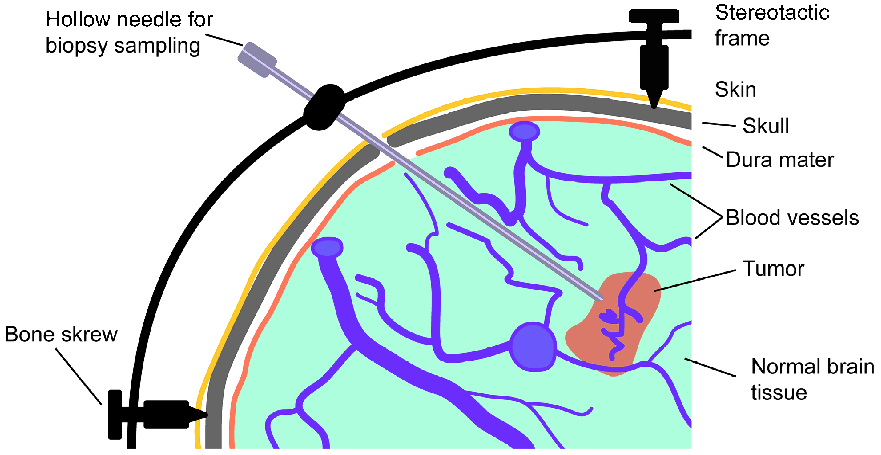
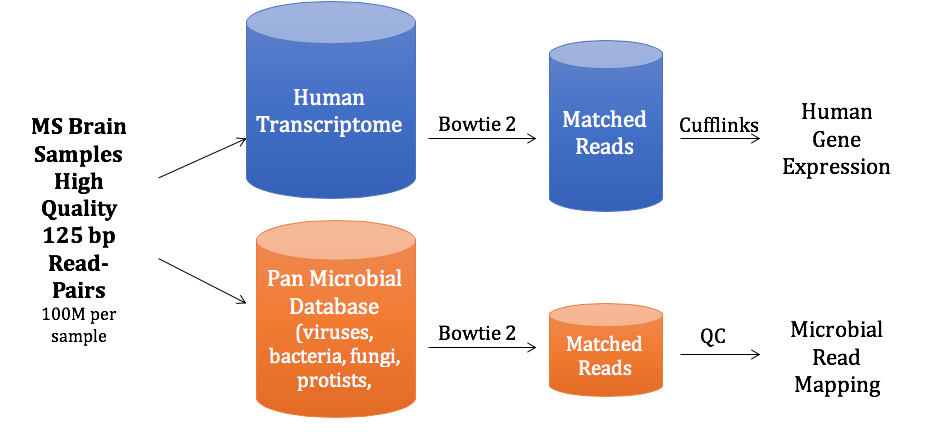
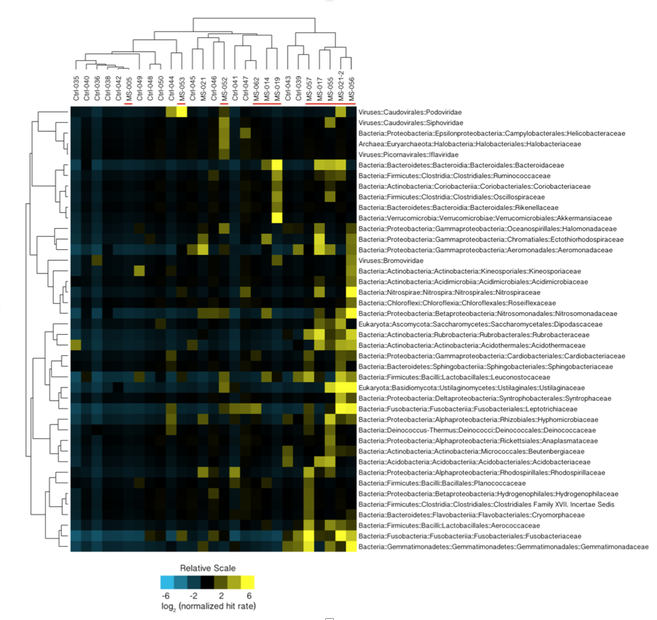
You’ve heard how soil samples can provide a deep look into the history of a landscape. Deep sequencing does the same with human tissue. This metagenomic testing provides unbiased “reads” of DNA or RNA sequences from the sample provided. It’s a complex process as you might guess, given the complexity of our human biosystems. When I tried to explain my research to my teenage son, he said it seemed like a crap shoot trying to find the cause of MS. I said, “It’s more like a wet spaghetti test: You throw your data up against the sequencing wall and see if it sticks.” Deep sequencing uses algorithms to splice together parts of RNA into whole strands called “paired-end” reads. As sequencing technology has improved, the reads have gotten longer and more precise. Here's a helpful illustration from Illumina's website: Why study RNA and not DNA? Simple. DNA is stable, RNA less so. DNA carries the genetic code so it must be stable and long lasting. RNA is made for a temporary purpose—messages, ribosomes, and other stuff—then it degrades and goes away. DNA is everywhere including the walls and ceiling of your house, on office machines, and even floating around in the air. RNA exists only within DNA and thus has less risk of contamination. So the goal of our sequencing studies is to see what microbes might be inhabiting diseased brain tissue from patients with MS. That is, if MS is related to one or more microbes—viruses, bacteria, fungi, protozoa, and/or archaeans—those microbes may very well be found at the site of the disease, the demyelinated areas within the brain. Now MS patients do not routinely have brain biopsies. MS can usually be diagnosed based on neurologic symptoms, brain MRI findings, and, sometimes, spinal fluid tests. But occasionally, doctors dispute a patient’s diagnosis. For instance, one University of Utah patient with pre-existing MS suddenly suffered a drastic downturn, such that she could no longer speak or walk. Her MRI showed a single, new, very large periventricular lesion (affecting a ventricle of her brain). Was it brain cancer or a glioblastoma (a malignant nervous system tumor)? Only a biopsy would tell. Her brain biopsy showed no cancer, just “primary demyelination”—the hallmark of MS. We know this because we sequenced her brain biopsy. Happily, she revived quickly and over the next few months, she returned to her previous quite functional state. We have no idea why. The procedure for a stereotactic brain biopsy is depicted. Each MS patient in our study had one of these procedures. Over the years, our Microbes in MS research group at the University of Utah has collected about 20 such cases where a patient’s brain was biopsied looking for cancer, and the result was “primary demyelination.” We decided to put these samples to the spaghetti test and map the reads looking for microbes. Our first deep sequencing study, in 2012, produced 36 base pair (bp) single end reads, and about 10 million reads per sample. These reads represented RNA extracted from diseased brain tissue from several patients who died with MS, and similar normal brain tissue from control patients without any brain disease. For the first time anywhere, the testing identified the virus GBV-C in one patient’s brain tissue. Unfortunately, no other brain specimens we studied contained this virus, so we do not believe that GBV-C has anything to do with MS. That’s the nature of research, try and fail and fail and try again. A word about controls. An ophthalmology professor and friend once said, “Controls are everything.” That was some good advice. Ideal controls for our study would be the brain tissues from patients with no neurologic diseases who were about the same age and sex as the MS patients. But we do not live in an ideal world. Normal patients don’t get brain biopsies. Normal tissues are available from deceased persons, in fact they are readily available, but RNA degrades quickly after death, as it should. We had to turn elsewhere. Our neuropathologist had a suggestion: Why not use epilepsy brain samples? Some epilepsy patients have seizures that only brain surgery can stop, so brain biopsies are readily available. But there is a complicating factor. The seizure surgery requires two steps: first, mapping the part of the brain causing the seizures by placing electrodes there, and second, the brain biopsy itself. This two-part surgery can allow for more microbial contamination, so more microbes (and more microbial sequences) are expected in the epilepsy control specimens compared to the MS group who had only one surgery—their biopsy. Even so, using epilepsy brain biopsies as controls turned out to be a great idea . . . or at least the best idea we could come up with. Monks in the machine. The output of this sequencing process is paired-end sequences. Lots of them. So many, in fact, that it would take a castle full of monks thousands of years to do all the math. Fortunately, much to the relief of the monks, we have supercomputers to do the comparisons of about 150 million reads per sample, more than a billion overall, per patient. Sequencing Output and Analysis Scheme We compared 12 “primary demyelination” (i.e. MS) samples with 15 epilepsy controls. One of the first things we noticed was that the MS samples were “dirtier” than the controls. That is, they had more microbial sequence mappings (128 per million sequences) than the controls did (77 per million). Much to our surprise. And scientific delight. What were all those microbes up to? We then used a technique called the False Discovery Rate (FDR) to see what microbes might be enriched in the MS brain specimens. This required some normalization of the sequencing data so we could compare specimens directly. Since we don’t believe that MS is caused by any single microbe, we had each MS specimen compared to the set of controls. The “cluster” figure from our findings on the “heat map,” below, shows overrepresented (significantly higher expression) microbial families in each row. Each column represents a different sample: either MS (red bars) or epilepsy controls. The computer application (Cluster 3.0) arranges the microbial families and the samples by their mathematical relatedness, allowing us to see what goes together. Yellow is hot (overexpression), blue is cold (under-expression), and black is in the middle (median). Notice the cluster of 4-5 MS samples on the right side of the figure. All those yellow boxes show that these specimens were particularly “hot” with an increased number of sequences derived from multiple microbes. Our Microbes in MS "HEAT MAP" Results—Ten of the 12 MS brain samples we studied had at least some microbial sequence enrichment at the genus level. Once we stopped smiling, and jumping around the lab, we had the machine monks do some additional filtering of the data, leading to a list of MS candidate microbes. Additional filtering led to a manageable candidate list with 29 microbial members. Not all of these MS candidate microbes are likely to be real contenders but some are worth discussing. For instance, Fusobacterium, Aggregatibacter, and Bacteroides are well known human pathogens. All three can get into the bloodstream; this is called bacteremia. Another contender, Atopobium, raised our eyebrows and our pulses. It’s part of the normal vaginal anaerobic flora and is not a known pathogen—but having vaginal bacteria appear in the brain—we know not how—could explain a lot since two-thirds to three-fourths of MS patients are female. (Neuroscientists recently found gut bacteria inhabiting the cells of healthy brains, a preliminary study which one UCLA doctor called “mind-blowing.”) Do we believe all of these MS microbial candidates are real culprits in causing MS? No. It’s hard to imagine how Ustilago, the corn smut fungus, could get into people and cause disease. Likewise, Nitrosospira and two related genera are common environmental organisms. We used the controls and fancy statistical comparisons to filter out spurious findings but there were millions of sequences compared to thousands of microbial taxa. And so the research team, and the monks, have to keep an open mind. If some spurious microbes found their way into the data, we will sleuth them out. Stay tuned for the list of our winning microbial candidates in my next blog, and for a future blog on antibacterial immunohistochemistry—another way to gain new and supportive research evidence to help find the cause of MS. Shutterstock ID: 777790381
0 Comments
Your comment will be posted after it is approved.
Leave a Reply. |
AuthorDr. John Kriesel is Associate Professor of Infectious Diseases at the University of Utah School of Medicine. He began this blog to raise awareness and generate discussion about the possible causes of multiple sclerosis. Archives
March 2021
Categories |
 RSS Feed
RSS Feed